Data Sources
DOHMH Rodent Inspection Data
The rodent
inspection data contains the information on rat sightings and
intervention visits in NYC from 1918 to 2022, which is managed by New
York City Department of Health and Mental Hygiene, Division of
Environmental Health. The data source is the Veterinary, Rodent and
Vector Surveillance System (VRVSS). It is also available on the Rat
Information Portal, which is a web-based mapping application where
users can view and map rat inspection and intervention data. Users can
search results from Health Department inspections for rats at the level
of individual properties, and view neighborhood maps.
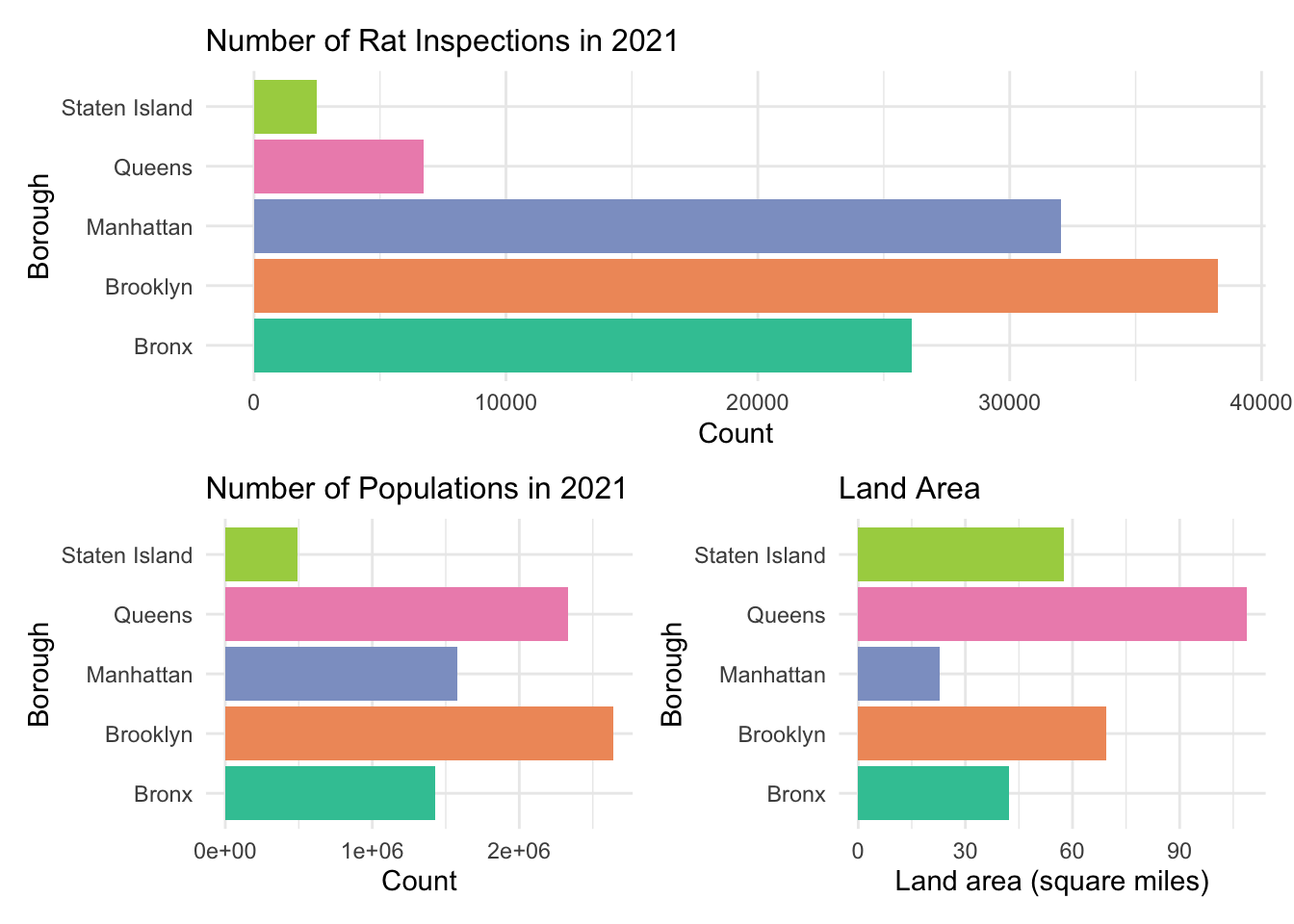
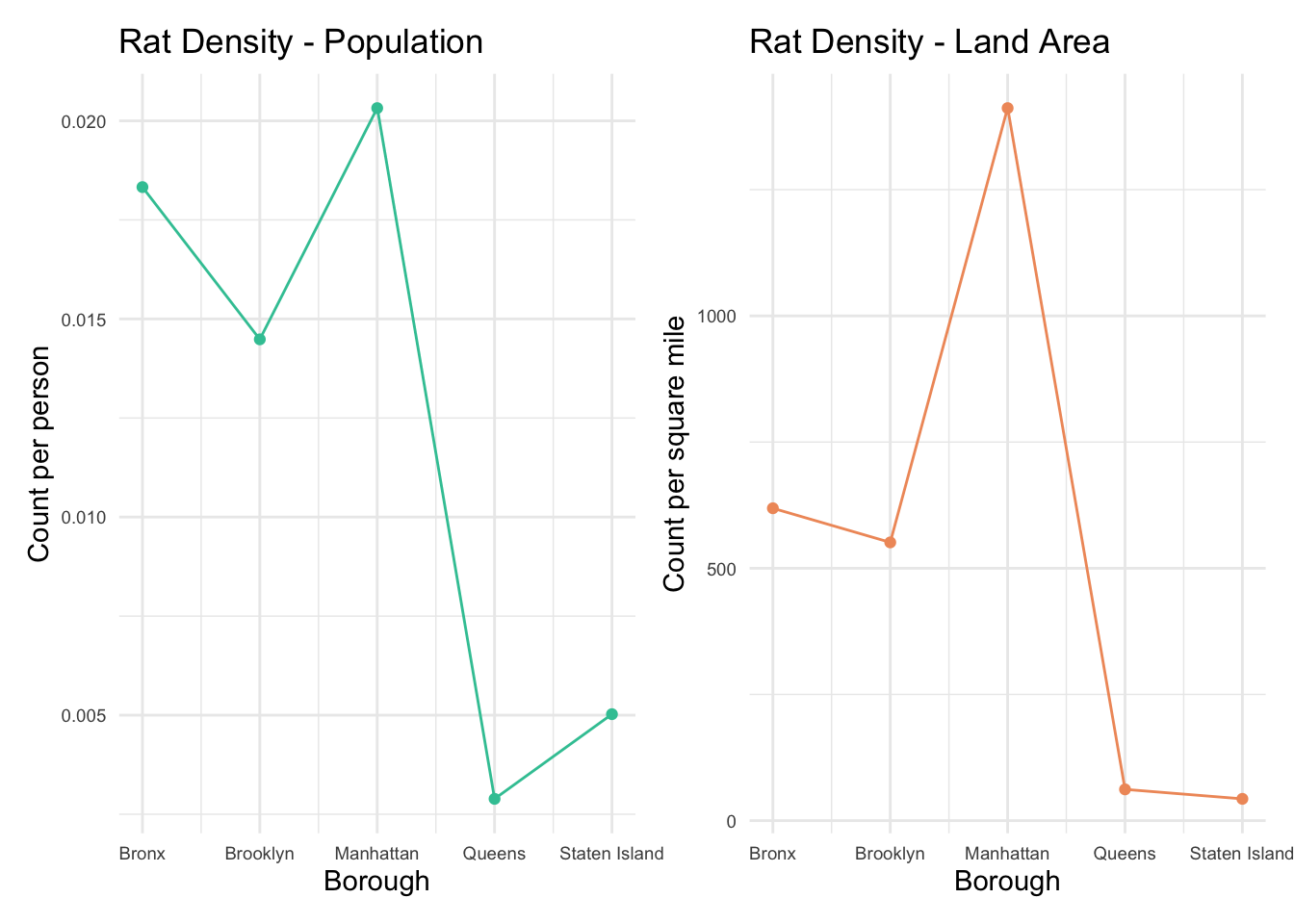
What should to be mentioned is that most of the inspection is due to
the complaints from the general public. Thus, if a property/taxlot does
not appear in the file, which does not indicate an absence of rats -
rather just that it has not been inspected. Similarly, neighborhoods
with higher numbers properties with active rat signs may not actually
have higher rat populations but simply have more inspections.
NYC Daily Weather Data
The NYC
daily weather data from the National Oceanic and Atmospheric
Association (NOAA) of the National Centers for Environmental Information
(NCEI), consists of the NYC daily climate observations from the Central
Park station (id: USW00094728), which is integrated by the GHCN (Global
Historical Climatology Network)-Daily database. The data is also
available to be retrieved using R functions from the rnoaa
package.
Global Historical Climate Network includes daily land surface
observations from over 100,000 stations in 180 countries and
territories. The GHCN-Daily was developed to meet the needs of climate
analysis and monitoring studies that require data at a sub-monthly time
resolution (e.g., assessments of the frequency of heavy rainfall, heat
wave duration, etc.). NCEI provides numerous daily variables, including
maximum and minimum temperature, total daily precipitation, snowfall,
and snow depth; however, about one half of the stations report
precipitation only. Both the record length and period of record vary by
station and cover intervals ranging from less than a year to more than
175 years.
NYC COVID-19 Daily Case Count Data
The NYC
COVID-19 daily case count data, provided by New York City Department
of Health and Mental Hygiene (DOHMH), represents citywide and
borough-specific daily counts of COVID-19 confirmed cases and
COVID-related hospitalizations and confirmed and probable deaths among
New York City residents. The case counts are aggregated by date of
diagnosis (i.e., date of specimen collection), and the hospitalization
cases areaggregated by date of admission, and death cases are aggregated
by date of death.
Data collection is since February 29, 2020, which is the date that
the Health Department classifies as the start of the COVID-19 outbreak
in NYC as it was the date of the first laboratory-confirmed COVID-19
case. Data on confirmed cases were passively reported to the NYC Health
Department by hospital, commercial, and public health laboratories. In
March, April, and early May, the NYC Health Department had discouraged
people with mild and moderate symptoms from being tested, so our data
primarily represent people with more severe illness. Until mid-May,
patients with more severe COVID-19 illness were more likely to have been
tested and included in these data. Data on hospitalizations for
confirmed COVID-19 cases were obtained from direct remote access to
electronic health record systems, regional health information
organization (RHIOs), and NYC Health and Hospital information, as well
as matching to syndromic surveillance. Deaths were confirmed by the New
York City Office of the Chief Medical Examiner and the Health
Department’s Bureau of Vital Statistics. Data counts are underestimates.
This dataset has been available to the public since May 19, 2020, and is
updated on a daily basis.
These data can be used to:
- Identify temporal trends in the number of persons diagnosed with
COVID-19, citywide and by borough.
- Identify temporal trends in the numbers of COVID-19-related
hospitalizations and deaths, citywide and by borough.
NYC Borough Population Data
The NYC
borough population data, provided by City Population, consists of
the population of the boroughs of New York City according to the U.S.
Census Bureau results. Additional information about the population
structure including gender, age groups, age distribution, race,
ethnicity can be also found.
Data Cleaning
Due to the large size of our final project datasets, another repository
is created for storing tidied data. The codes below describes the steps
we took to collate and merge dataset we used in the following
exploratory and statistical analysis. Moreover, the detailed data
preprocessing procedures can be find here.
# deal with rat inspection data
url_rat = "https://data.cityofnewyork.us/OData.svc/p937-wjvj"
rat = read.socrata(url_rat) %>%
janitor::clean_names()
rat_tidy = rat %>%
select(inspection_type, bbl, zip_code, street_name, latitude, longitude, borough, result, inspection_date, approved_date) %>%
drop_na() %>%
mutate(boro_code = substr(bbl, 1, 1),
block = substr(bbl, 2, 6),
lot = substr(bbl, 7, 10)) %>%
select(inspection_type, boro_code, block, lot, zip_code, street_name, latitude, longitude, borough, result, inspection_date, approved_date) %>%
separate(inspection_date, c("inspection_date", "inspection_time"), " ") %>%
separate(inspection_date, c("inspection_year", "inspection_month", "inspection_day"), "-") %>%
separate(approved_date, c("approved_date", "approved_time"), " ") %>%
separate(approved_date, c("approved_year", "approved_month", "approved_day"), "-") %>%
mutate(inspection_year = as.integer(inspection_year),
inspection_month = as.integer(inspection_month),
inspection_day = as.integer(inspection_day)) %>%
mutate(approved_year = as.integer(approved_year),
approved_month = as.integer(approved_month),
approved_day = as.integer(approved_day)) %>%
relocate(inspection_year, .before = "inspection_month") %>%
relocate(approved_year, .before = "approved_month") %>%
arrange(inspection_year, inspection_month) %>%
mutate(inspection_month = month.abb[inspection_month],
approved_month = month.abb[approved_month]) %>%
filter(inspection_year >= 2012 & inspection_year <= 2021)
# deal with weather data
nycstationsid = ghcnd_stations() %>%
filter(id == "USW00094728") %>%
distinct(id)
nyc_weather = meteo_pull_monitors(nycstationsid$id,
date_min = "2012-01-01",
date_max = "2021-12-31",
var = c("PRCP", "SNOW", "SNWD", "TMAX", "TMIN"))
nyc_weather_tidy = nyc_weather %>%
janitor::clean_names() %>%
separate(date, into = c("year", "month", "day")) %>%
mutate(year = as.numeric(year),
month = month.abb[as.numeric(month)],
day = as.numeric(day)) %>%
mutate(prcp = prcp/10,
tmax = tmax/10,
tmin = tmin/10)
# deal with covid data
url_covid = "https://data.cityofnewyork.us/OData.svc/rc75-m7u3"
covid = read.socrata(url_covid) %>%
janitor::clean_names()
covid_tidy = covid %>%
rename(date = date_of_interest) %>%
select(date, contains("case_count")) %>%
select(-contains(c("probable_case_count", "case_count_7day_avg", "all_case_count_7day_avg"))) %>%
separate(date, into = c("year", "month", "day")) %>%
mutate(year = as.numeric(year),
month = month.abb[as.numeric(month)],
day = as.numeric(day)) %>%
pivot_longer(
cols = bx_case_count:si_case_count,
names_to = "borough",
values_to = "borough_case_count"
) %>%
mutate(borough = gsub("_case_count", "", borough)) %>%
mutate(borough = dplyr::recode(borough, "bx" = "Bronx","bk" = "Brooklyn","mn" = "Manhattan","si" = "Staten Island","qn" = "Queens")) %>%
relocate(case_count, .after = borough_case_count) %>%
rename(total_case_count = case_count)
# merge the above dataframe with covid info.
rat_weather_covid = rat_tidy %>%
filter(inspection_year %in% c(2020,2021)) %>%
merge(nyc_weather_tidy, by.x = c("inspection_year","inspection_month","inspection_day"), by.y = c("year","month","day")) %>%
select(-id) %>%
merge(covid_tidy, by.x = c("inspection_year","inspection_month","inspection_day","borough"), by.y = c("year","month","day","borough"))
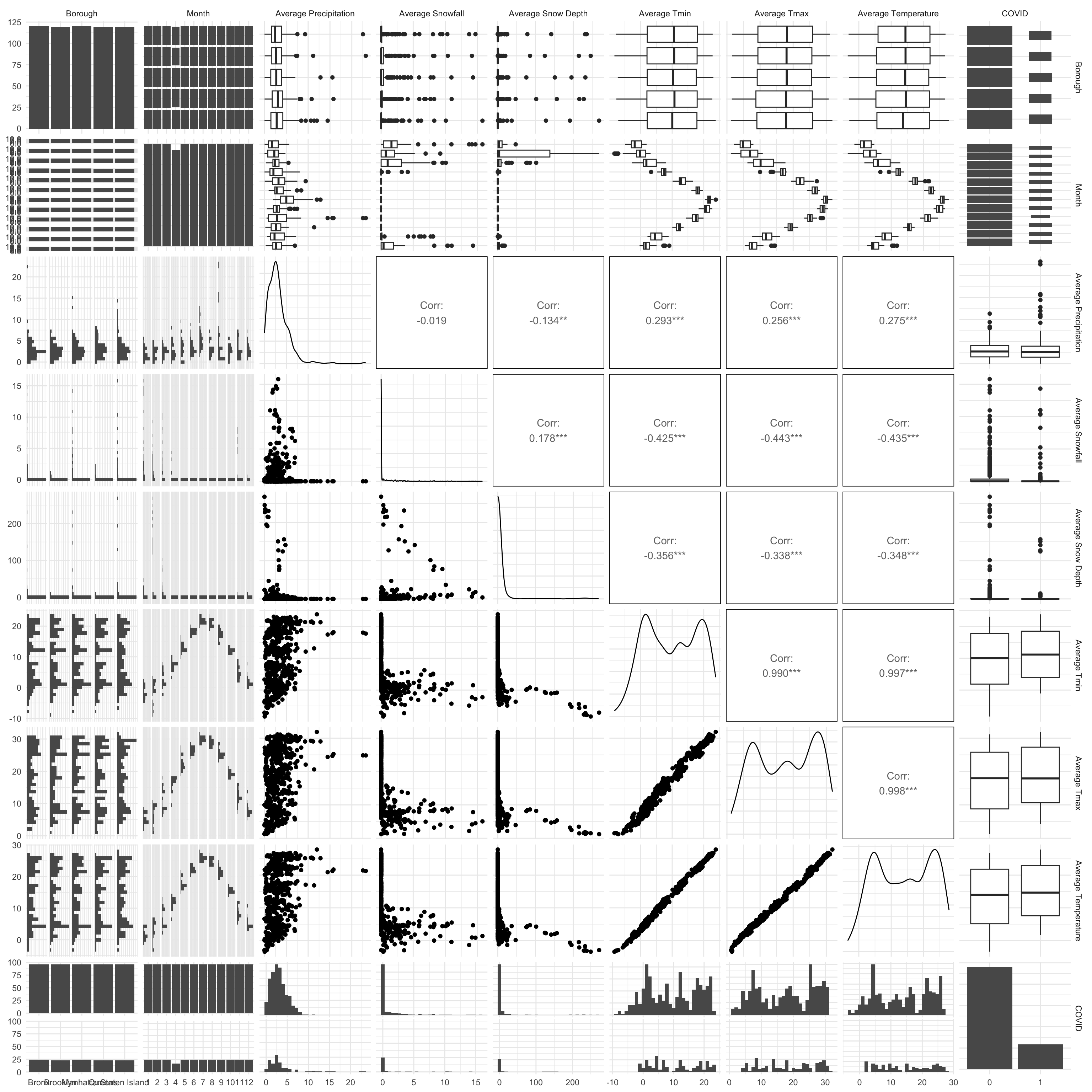
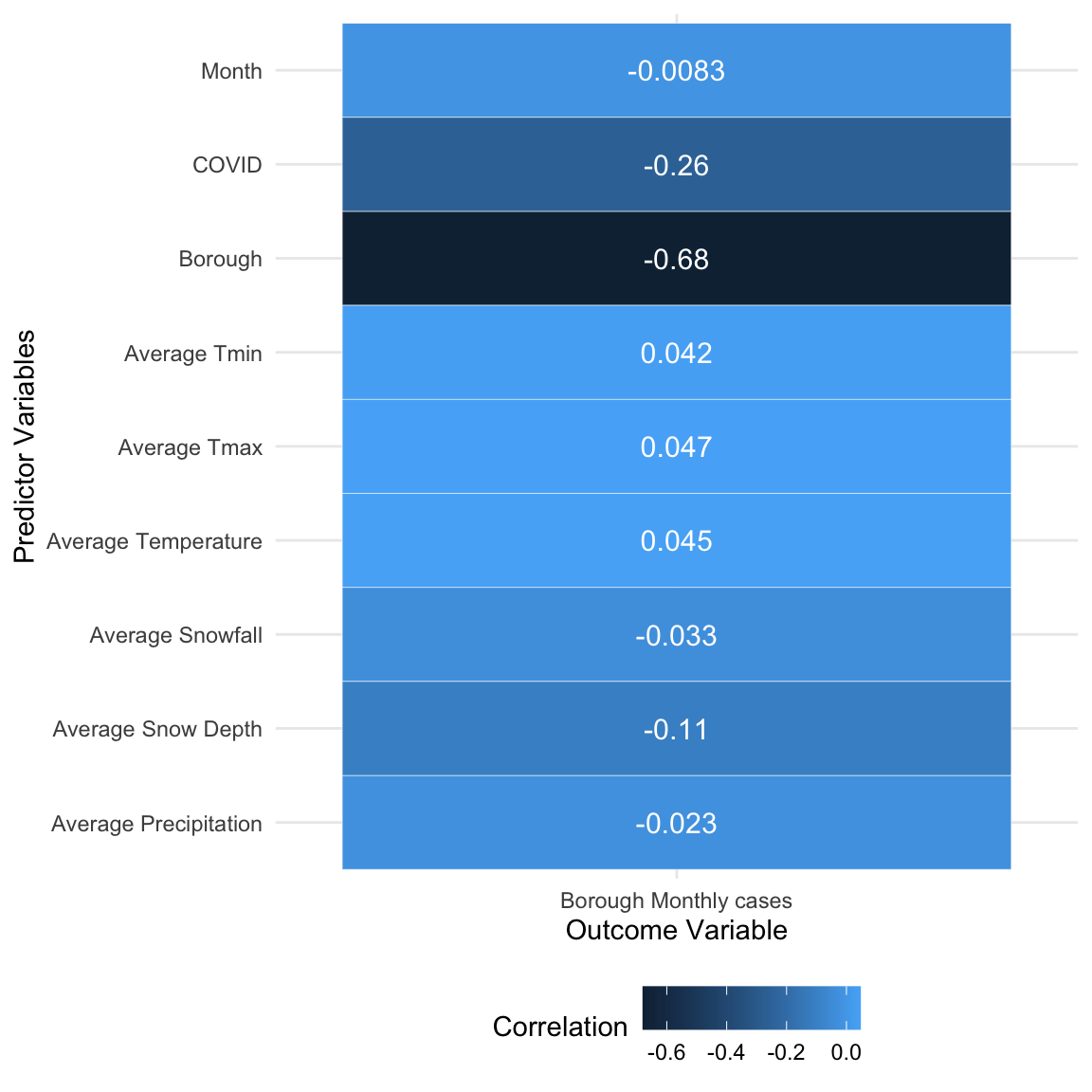
After cleaning the datasets mentioned above, our tidied and
aggregated data has a total of 1,679,675 rows, one for each rat
inspection record, and is the basis of our exploratory and statistical
analysis. Altogether 24 variables are selected as meaningful and
valuable. The specific variable names and their corresponding
explanation are listed below:
inspection_year: year of the inspectioninspection_month: month of the inspectioninspection_day: day of the inspectionboro_code: code assigned to the NYC boroughblock: block number for the inspected taxlot; block
numbers repeat in different boroughslot: lot number for the inspected taxlot (Notes: lot
numbers can repeat in different blocks)zip_code: postal zipcode of the taxlot that was
inspectedstreet_name:latitude: latitude in decimal degrees of the inspected
taxlot (WGS 1984)longitude: longitude in decimal degrees of the
inspected taxlot (WGS 1984)borough: name of the NYC boroughresult: result of the inspection including Active Rat
Signs (ARS) and Problem Conditionsinspection_time: time of the inspection.approved_year: year of the inspection approved by
supervisor at DOHMHapproved_month: month of the inspection approved by
supervisor at DOHMHapproved_day: day of the inspection approved by
supervisor at DOHMHapproved_time: time of the inspection approved by
supervisor at DOHMHprcp: precipitation (mm)snow: snowfall (mm)snwd: snow depth (mm)tmax: maximum temperature (°C)tmin: minimum temperature (°C)borough_case_count: count of patients tested who were
confirmed to be COVID-19 cases on date_of_interest in
borough_of_interesttotal_case_count: total count of patients tested who
were confirmed to be COVID-19 cases on date_of_interest in NYC